| xiv) | Now we need to tweak some of the information we previously entered. |
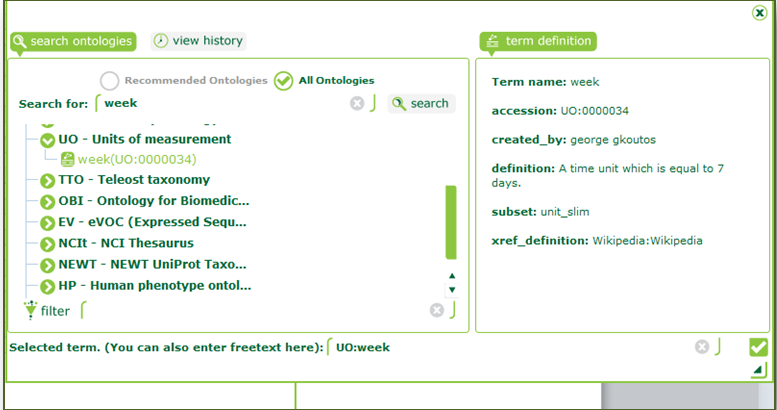
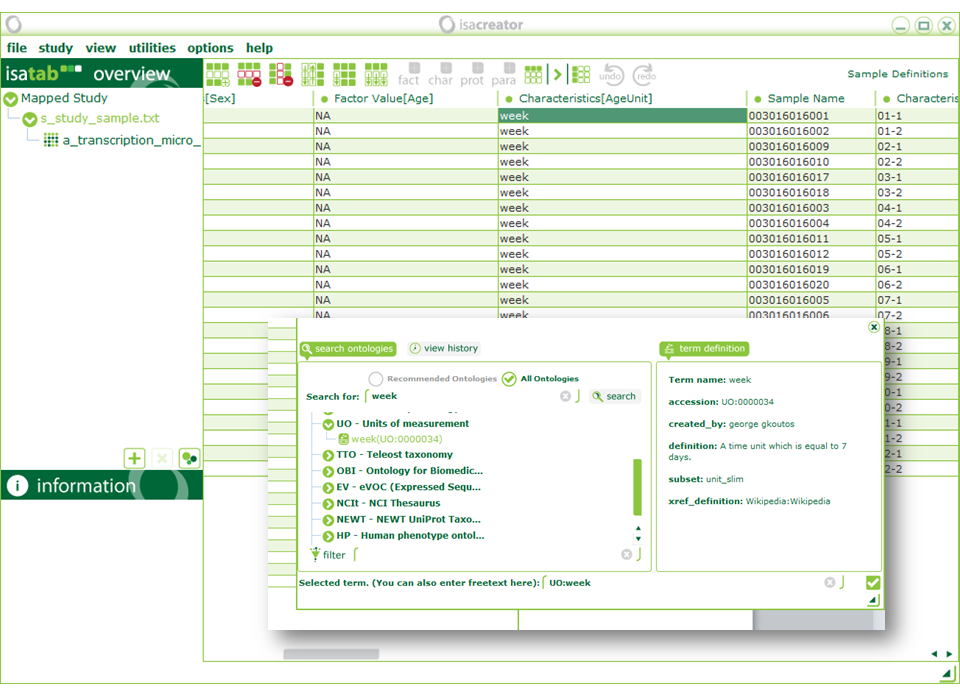
| Click on “s_study_sample.txt” on the left menu and scroll right to “Characteristics[AgeUnit]”. Double-click on the first entry and search for “week”. Select the term “UO:week” from the Units of Measurement ontology. |
| Ontology harmonisation is an important step towards interoperability of data. Imagine a case where “week” was indicated by different researchers as “w”, “WK”, etc., making it ambiguous. Linking to an ontology reference gives our term a standard name and a definition. |
Click on the “copy column downwards” button to replace the text we previously entered with the ontology term. Identifiers for the SEURAT Gold compounds are available in the gold compounds database (wiki.toxbank.net). If the general search term (e.g. human) produces too many matches you can use the more specialized terms from the ontology below. When doing your own files try to use the same onlogies as below for similar terms but the use of any ontology, as opposed to free text, will still enhance the file.
| • | We can similarly replace “Human” (Characteristics[Organism]) with “NCBITaxon:Homo sapiens” (search for “Homo sapiens” and select “obo:NCBITaxon_9606”) |
| • | “liver” (Characteristics[Organ]) with “OBI:liver” (select “obo:UBERON_0002107”) |
| • | “acetaminophen” (Factor Value[Compound]) with “CHEBI:paracetamol” Exact match “paracetamol(CHEBI:46195)” |
| • | “micromolar” (Characterictics[DoseUnit]) with “UO:micromolar” Exact match “micromolar(UO:0000064)” |
| • | “hour” (Characteristis[SampleTimePointUnit]) with “UO:hour” Exact match “hour(UO:0000032)” |
| • | “percent” (Unit) with “UO:percent” Exact match “percent(UO:0000187)” |
| Additional Information: The ISA-Tab suite of tools includes ISA2RDF, developed by ToxBank. ISA-Tab files can be converted to RDF (Resource Description Framework) format files, the standard file format of the semantic web. Terms from various Ontologies (or the SEURAT-1/ToxBank keyword hierarchy) will connect your data directly to the “semantic cloud”. Unambiguously defined terms can be used as keys and identifiers to connect your experiment to the entire world’s biomedical and toxicological data. These kinds of connections will be crucial for “Integrated Data Analysis” phase of the SEURAT-1 project. |
| xv) |
We also need to make the following adjustments: |
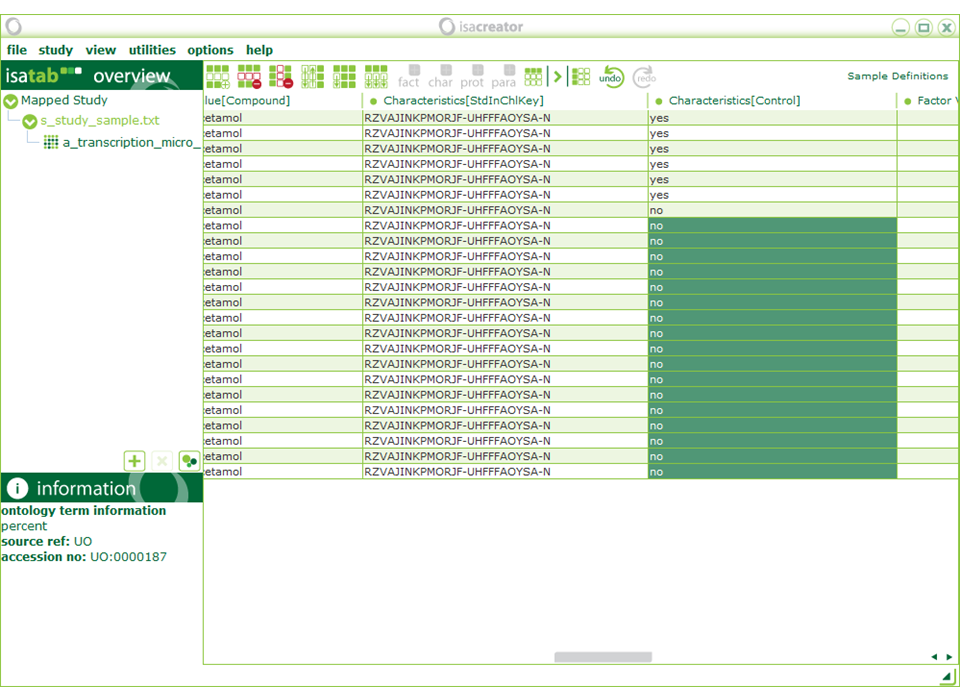
| “Characteristics[Control]” is a yes/no field so we need to replace “Control” with “yes” and everything else with “no” (we’ve recorded the different levels under “Characteristics[TreatmentGroup]” so we won’t lose this information). When you start typing a ontology search window pops up. You can enter text by typing the first entry (in the dialog box in the position: “you can also enter free text here”), then dragging it across the columns that should be the same, and selecting “Autofill” from the menu that pops-up. |
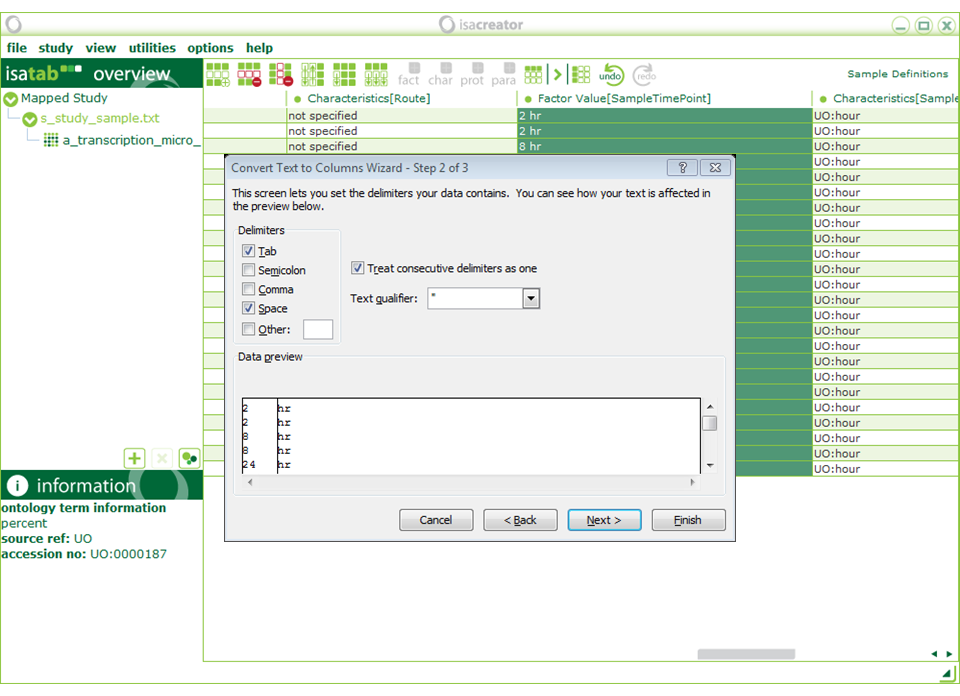
From “Factor Value[SampleTimePoint]” we need to remove the “hr” in order to make it a numeric field (we’ve recorded “UO:hour” in the Characteristics [SampleTimePointUnit]”). You can do this by hand in ISAcreator, or by copying and pasting it in Excel and then using functions to automatically extract the numerical part from the string (e.g. The “convert text to columns wizard” in the next screen shot). Using Excel can be convenient if the number of samples is very large. Otherwise doing it in ISAcreator might be easier.
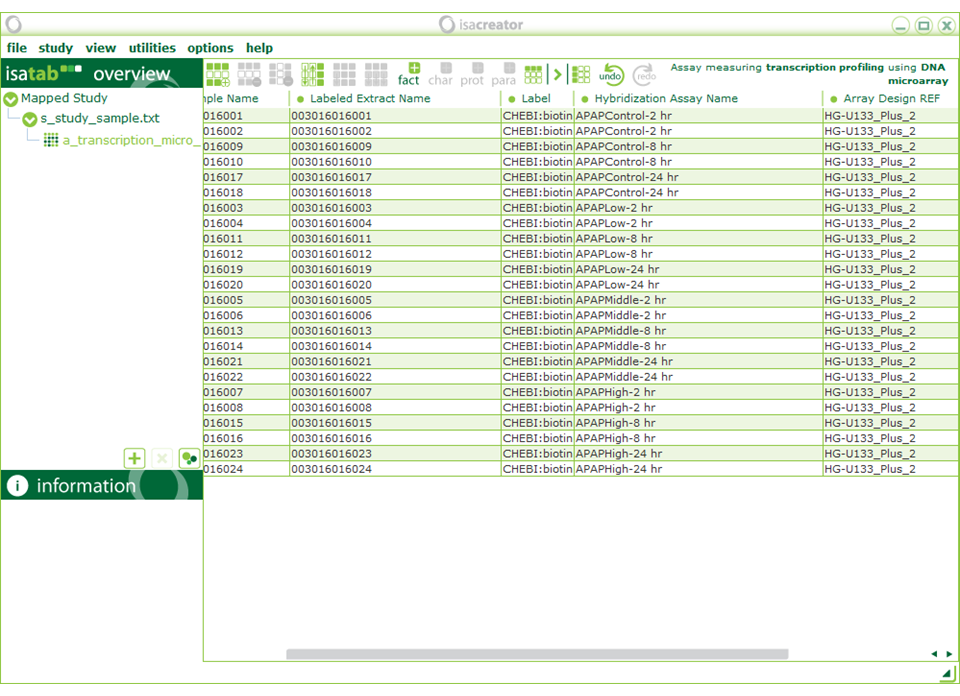
On the assay level (“a_transcription_micro” on the left menu), you can replace “biotin” (Label) with “CHEBI:biotin” (exact match “biotin(CHEBI:15956)”)
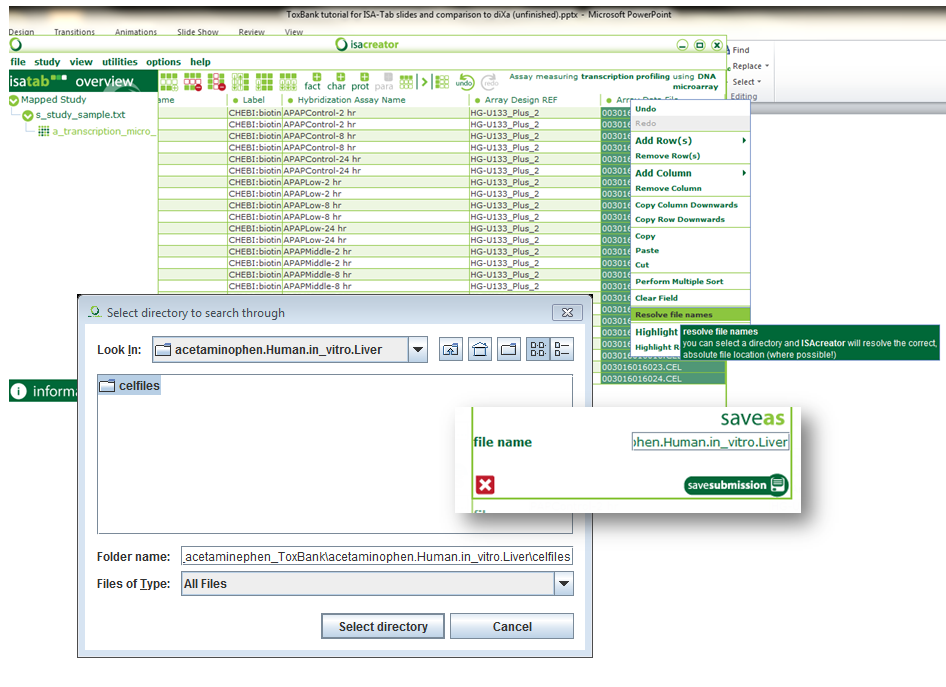
| xvi) | Finally, select the “Array Data File” column, right click and “resolve filenames”. Browse to the directory that contains your CEL files and “Select directory”. You now should have the full path to your CEL files be filled in. |
| xvii) | Congratulations! You have successfully created a standardised SEURAT-1/ToxBank ISA-tab archive. Click on File -> Save and give “acetaminophen.Human.in_vitro.Liver” as the folder name. |
| TIP: To be sure everything is in order you can validate your archive in File -> Validate ISAtab. |
| xviii) |
You can now save your ISA-tab investigation as a zip archive on your Desktop. Simply select save/save as, and then you can continue working on them. By default the files will be saved into the folder: …\ ISAcreator.SEURAT\isa-tab files. |
| Go to File -> Create ISArchive. |
A file named as “acetaminophen.Human.in_vitro.Liver_archive.zip” should appear on your Desktop.
This archive contains your study annotations in ISA-tab format, as well as, all the CEL files that go along with it. You can submit this file directly to the ToxBank Data Warehouse using the web interface.
For any questions regarding this tutorial, please email ToxBank Development List: support@lists.toxbank.net with “ISAtab tutorial” in the subject header.
Proceed to: